gobics.de [ Mario Stanke ]
University of Göttingen
- Faculty of
Biology - Institute
of Microbiology and Genetics - Department
of Bioinformatics
Research Topics:
Gene Prediction:
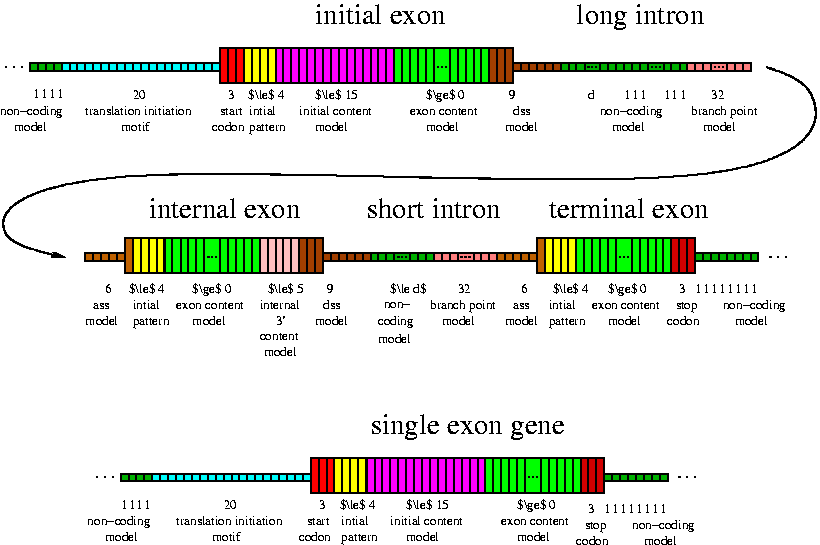
We are developping a gene prediction software that finds protein coding genes
in eukaryotic DNA. The program, which is called AUGUSTUS,
is based on a Semi-Markov linear-chain Conditional Random Field.
This model probabilistically models the base composition
of coding regions, untranslated regions, introns and intergenic regions, the donor and acceptor splice site, the
branch point region, the translation start, the lengths of exons and introns, the number of exons per gene and signals at the transcription end and start.
We have also developped a method for incorporating extrinsic information about the gene structure
in a way that allows for errors in the extrinsic information.
Related subtopics are:
- RNA-Seq incorporation: Effective use of Next Genration Sequencing (NGS) sequence data for gene finding.
- Protein-family based gene prediction: Use the knowledge about a particular protein family in order to precisely identify the structure of new members of the family in
naked genomes.
- Proteogenomics: Use of of Peptides from MS/MS to correct structural genome annotation.
- We are exploring methods with Mladen Tzvetkov for detecting whether polymorphisms
or mutations in the human genome can influence splicing.
Detecting Recombination in Viruses:
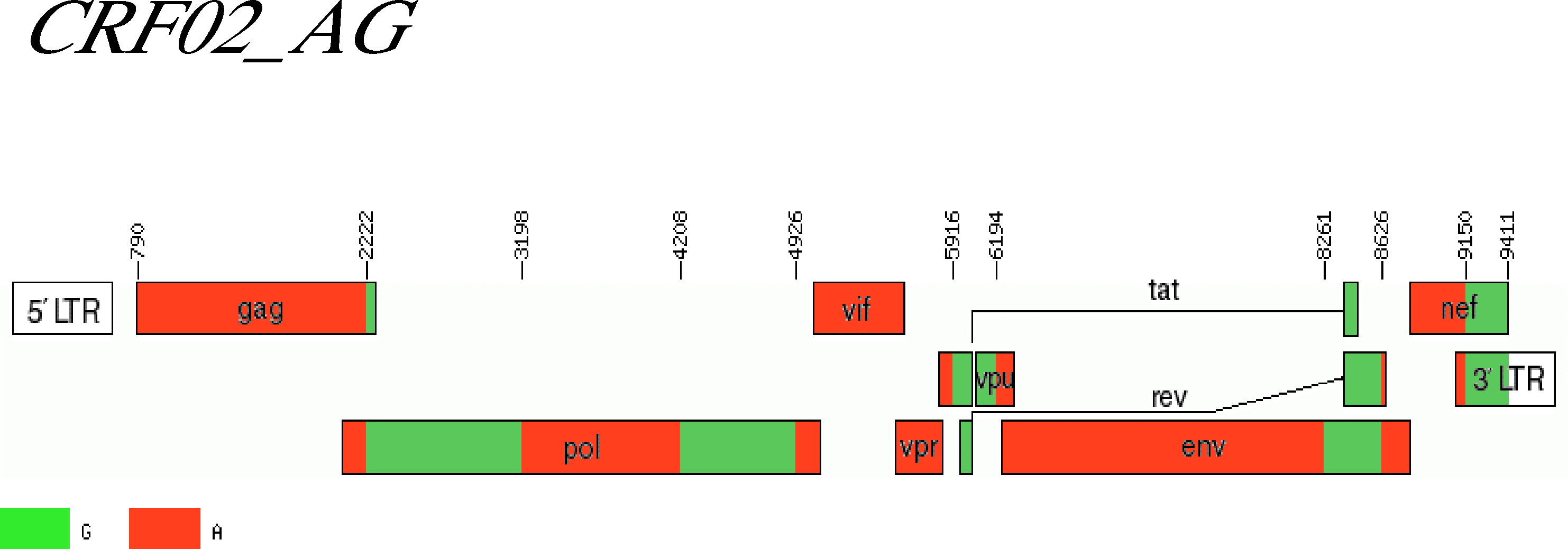
We developped a Hidden-Markov-Model (HMM) that is a generalization of profile HMMs, that are commonly used for modelling protein sequence families.
Our model is called a jumping profile HMM ans was programmed by Anne. It models a collection (superfamiliy) of sequence families and allows that a given sequence is locally similar to different
families in the collection. This model is applied in order to detect HIV sequences that are a recombination of HIV sequences from different subtypes of HIV.
Such recombinations are frequent in HIV and so-called Circulating Recombinant Forms play a significant role in the global pandemic.
We are collaborating with colleagues from the Los Alamos National Laboratory on HIV recombination.
Inferring the Genealogy of a Sequence Set in the Presence of Recombination:
Based on our experience with recombination in HIV, we aim at developing a general tool that predicts for a set of sequences its "history" like in a phylogenetic tree.
However the new method should also predict when there has been recombination in the past, and the ancestors of which current taxa have been involved. This work is chiefly done
by Ingo.
Detecting Protein-Protein Interaction Sites:
Together with Stephan Waack we are exporing methods based on Conditional Random Fields for predicting, which
resudiues in a protein belong to the interface and which residues don't.

