Definition
Metagenomics is "the genomic analysis of microorgansims by direct extraction and cloning of DNA from an assemblage of microorganisms" (Handelsman, 2004).
Experimental approaches to Metagenomics
Techniques of Metagenomics are generally used to explore the properties of microorganisms without prior cultivation. Although still a novel field of research, Metagenomics has already developed different branches, which contain mainly the following focuses:
- Discovery of new proteins by functional screening techniques (e.g. screening for an enzyme that is able to degrade a particular toxic compound),
- analysis of microbioal diversity in a habitat (e.g. by classical rRNA analysis or phylum estimation from sequence properties),
- and gene discovery by sequence analysis.
The last two approaches rely on metagenome sequencing and produce sequence data which can be analysed by computational methods.
Bioinformatics on Metagenomes
Sequenced metagenomes yield fragmented genomic data that is comprised from a mixture of anonymous microorganisms. Amoung others, bioinformatics can be used to
- sort the fragmented nucleotide fragments into 'bins' of different taxonomic/phylogenetic levels,
- assemble contigs from short sequence reads,
- predict genes on sequence reads (or assembled contigs),
- predict gene function,
- classify predicted proteins into families or other groups.
For the reason that metagenomics is a young field of research, the development of evaluation methods for algorithms that process metagenomic data is also a field of on ongoing research by itself.
An illustrated example for a metagenomic study
Many metagenome sequencing projects currently rely on 'shotgun sequencing', a method which is based on cloning and subsequent sequencing of genomic DNA. The single steps of such a metagenomic study are illustrated below.
Sampling
The first step of all metagenomic studies is the extraction of a sample from some environmental habitat. An environmental habitat could be
 |
 |
 |
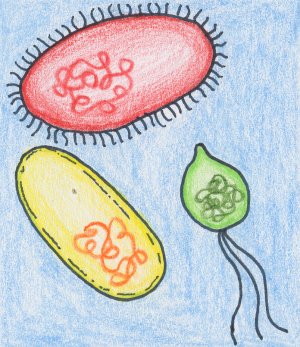
| The original environmental sample contains all material from the chosen environment, including the microorganisms living in there. The red, green and yellow thing symbolize microbes living in an environmental sample. |
 |
Cloning and Sequencing
The environmental samples are further on processed in a molecular biology laboratory. Some of the intermediate steps on the way to obtaining a sequenced metagenome are these:
 |
 |
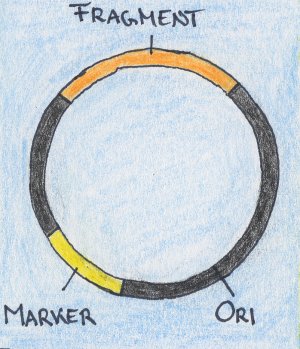 |
| DNA isolation: the genomic DNA of all microorganisms that are present in a sample is simultanously extracted. | Shearing: the isolated genomic DNA is broken into shorter fragments that can be cloned into plasmids (a vector for smaller insert size). Some DNA isolations methods already contain a shearing step by themselves. | Cloning: the DNA fragments are cloned into a vector. From here on, all steps are shown for one fragment, only. The vector contains an origin of replication which enables the host organsim to multiply the plasmid. It also contains a marker which assists in the selection of host organisms that have incorporated the plasmid |
 |
 |
 |
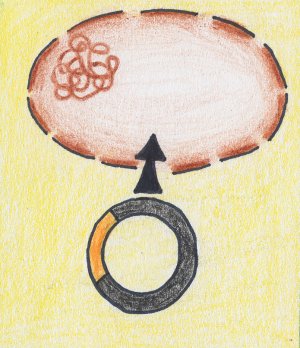
| Tranformation: constructs consisting of a plasmid with an insert are transformed into a host organism. | Multiplication: the host organism multiplies (and obtains) the transformed material. | Sequencing: after another step of DNA isolation which in this case isolates the multiplied plasmid material from the host organism, the single fragments can be sequenced by various methods. Usually, Sanger sequencing with fluorescence is applied. |
The resulting collection of DNA fragment sequences can be processed manually or by computational methods (see above).